Watching molecules move inside bacterial cells
For a long time, cells were seen as bags filled with randomly distributed molecules. We now know that cells with a nucleus contain mini-organs (organelles): compartments that contain the machinery to carry out specific tasks. Although bacterial cells have no such compartments, evidence suggests that even these cells are not homogeneous. Biochemists from the University of Groningen have now mapped diffusion rates inside a bacterial cell, which identified distinct regions. Their results were published in the journal Science Advances on 12 August.
‘We already know a lot about the compartmentalization of eukaryotic cells: cells with a nucleus, like the ones that make up our body. But we know much less about the structure of bacterial cytoplasm, mainly because bacteria are so much smaller,’ explains Bert Poolman, Professor of Biochemistry at the University of Groningen. ‘However, the resolution of optical microscopes is insufficient to see details in the structure of the cytoplasm in these small living organisms.’

Displacement
We do know which proteins and other large molecules that are present in bacterial cells. By using fluorescent probes, it is possible to measure how ‘crowded’ the cells are, which is important to understand the interactions between molecules and biochemical reactions inside cells. Levels of crowding can be estimated by measuring the mobility of molecules inside cells. Therefore, Poolman was very interested when he met a colleague at a conference who could measure the mobility of single molecules with much higher time resolution than had ever been reported before. ‘We realized that this could be a useful tool for investigating the structure of the bacterial cytoplasm.’
The technique is called Single-Molecule Displacement Mapping. ‘Basically, you attach a fluorescent probe to a molecule. The probe is activated by laser light and then read out using a second laser. The readout is done on a millisecond scale, which allows you to see how far the molecule has travelled between two readouts.’ By choosing the right conditions for activation, just one molecule at a time can be observed. By repeating the measurements tens of thousands of times, it is possible to determine diffusion coefficients at different locations inside a cell with high accuracy.
Hypothesis
Poolman and his team used fourteen different proteins to assess what dictates their diffusion in different parts of a cell. Poolman: ‘We chose proteins that differ in the number of interactions with other molecules, abundance and size.’ The hypothesis was that proteins with lots of interactions would move more slowly. ‘But this was not the case,’ explains Poolman. ‘Nor did we find any correlation between mobility and the number of copies of a particular molecule in the cell. The only factor that determined the diffusion rate turned out to be their effective size.’
Einstein
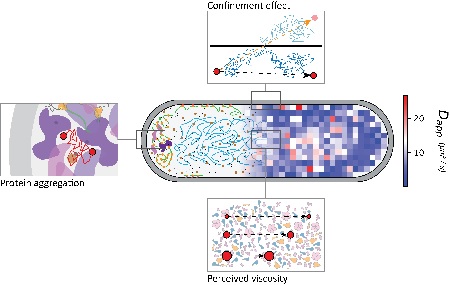
Interestingly, the diffusion coefficients did not conform to the classic Stokes-Einstein equation, which describes the random Brownian motion of molecules. ‘In a dilute solution, molecules experience the same viscosity regardless of their size. In the cytoplasm, the molecules followed an equation in which the viscosity depends on their size. Our idea is that the movement of larger molecules is hindered more by the presence of other molecules. Therefore, the cytoplasm feels more viscous to a larger molecule than a smaller one and, consequently, it moves slower.’
The diffusion coefficients of the fourteen proteins were also determined in different segments of the cell: the middle section, (about 60 percent of the cell volume) containing the cell’s DNA and the two poles (left and right) which each take up 20 percent of the volume. ‘Our observations show that diffusion coefficients are highest in the middle section. Interestingly, the extent of the mobility slowdown that we observed in the poles of a cell was far greater than what we expected to observe due to the geometry of these compartments,’ says Poolman. ‘This is probably because some extremely large molecular complexes, such as ribosomes during protein production, are pushed out from the middle section by the DNA towards the poles, making them more crowded. This would explain the hindered mobility that we observed.’
Optimized
The mobility of molecules inside a cell is not very high because the cytoplasm is highly crowded. Poolman: ‘Inside a cell, the diffusion coefficients are 10 to 100 times lower than in water.’ If the crowding were to increase by 20 to 30 percent, which happens when the amount of water in the environment changes, the molecules become so tightly packed that proteins are no longer capable of finding each other. This effectively stops biochemical reactions. ‘Apparently, the crowding of the cell is optimized for high reaction rates, at the edge of the point of diminishing return, where crowding is too high for molecules to meet effectively. We now have to figure out the physiology behind the differences in diffusion at the poles versus the middle of the cell.’
Reference: Wojciech M. Śmigiel1, Luca Mantovanelli, Dmitrii S. Linnik, Michiel Punter, Jakob Silberberg, Limin Xiang, Ke Xu and Bert Poolman: Protein diffusion in Escherichia coli cytoplasm scales with the mass of the complexes and is location dependent. Science Advances, 12 August 2022
| Last modified: | 28 November 2024 3.33 p.m. |
More news
-
20 December 2024
Three FSE researchers receive NWO M1 grant
Dr. Antonija Grubišić-Čabo, Dr. Robbert Havekes and Prof. Jan Komdeur receive an NWO M1 grant
-
19 December 2024
NWO ENW-XL million grants for RUG research projects
Four researchers from the Faculty of Science and Engineering (UG) receive NWO grants of 3 million euros for their research projects.
-
19 December 2024
Jacquelien Scherpen honoured with Hendrik W. Bode Lecture Prize 2025
For her achievements in the scientific development of control systems and engineering, Rector Jacquelien Scherpen has received the 2025 Hendrik W. Bode Lecture award from the IEEE Control Systems Society (CSS).

