The dance of DNAJB6b at atomistic resolution
Molecular chaperones are the guardians of protein structure. A particularly efficient one, DNAJB6b, ensures that unfolded proteins do not aggregate and cause neurodegenerative diseases. How exactly this chaperone does this is still unclear, partly because its structure is not fully known. Together with researchers from the UMCG, the Onck group has uncovered that DNAJB6b is a dynamic molecule interconverting between three different structural conformations, by performing massively-parallel atomistic molecular dynamics simulations. Their findings have been published on April 16th in the journal Nature Communications.
Proteins play a crucial role in our body and fulfill a wide range of functions. There are more than 100,000 different proteins, all of which have a different role. In most proteins, this role is directly dependent on their three-dimensional structure. However, if proteins are unfolded (i.e., do not have a 3D structure) or if they temporarily unfold, there is a lurking danger that they will clump together, leading to neurodegenerative diseases such as Huntington's, Alzheimer's, and Parkinson's. Fortunately, we also have a very efficient protein quality control system of molecular chaperones that prevents this process. In recent years it has been found that a chaperone called DNAJB6b does this very efficiently in various age-related diseases. How exactly this chaperone does this is still unclear, partly because the three-dimensional structure is unknown. In this work, the Zernike Institute researchers performed atomistic molecular dynamics simulations showing that these chaperones are not static proteins but continuously change their conformation.
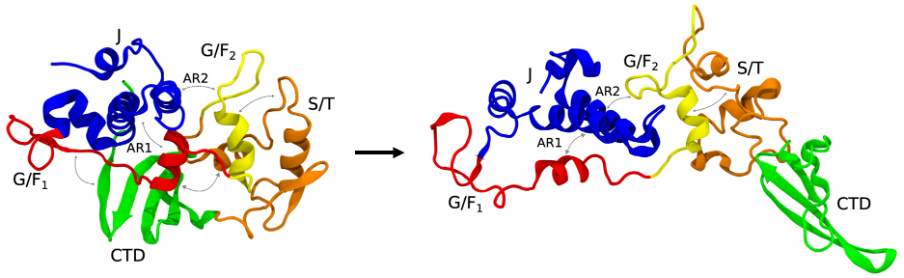
Microsecond-long all-atom explicit water molecular dynamics (MD) simulations were performed, revealing that DNAJB6b cycles through three different conformations (states): a closed, open, and extended state. DNAJB6b belongs to the canonical class B J-domain proteins, where the defining feature is the auto-inhibitory state. Autoinhibition is a self-regulatory mechanism in which the protein blocks its binding sites to prevent over-activation of the protein. In the case of DNAJB6b, this autoinhibition is achieved by blocking the active sites on helices II and III by another helix; helix V. The simulations found that although DNAJB6b cycles dynamically between different conformations, the autoinhibition always remains present. This indicates that the autoinhibition is only released if needed, e.g. in the presence of a substrate and HSP70.
Mutations in DNAJB6b have been linked to Limb-Girdle muscular dystrophy type D1 (LGMDD1), but how these mutations cause the disease is unknown. To explore this, they performed additional simulations and found that the native autoinhibition state is spontaneously released upon application of the LGMDD1 mutations (see animation). This means that the active binding sites are continuously exposed, even without a substrate, leading to over-activation of the chaperone system, potentially causing the pathogenesis of LGMDD1.
Reference: Adupa, V., Ustyantseva, E., Kampinga, H.H. and Onck, P.R., Tertiary structure and conformational dynamics of the anti-amyloidogenic chaperone DNAJB6b at atomistic resolution. Nat Commun 15, 3285 (2024). https://doi.org/10.1038/s41467-024-46587-z.
More news
-
15 September 2025
Successful visit to the UG by Rector of Institut Teknologi Bandung
